Pan-genome sequencing
Genome sequencing
Genome sequencing and assembly is the use of second-generation, third-generation and Hi-C sequencing technology, the use of bioinformatics methods to assemble and obtain high-quality genome reference sequences of species, and annotate the genome. At present, the characteristics of three-generation sequencing, which is long reading and can easily span most repeat sequence regions or heterozygous regions, make the genome assembly more complete and accurate. Genome sequencing can not only obtain the whole genome sequence map of species, but also lay a foundation for the subsequent study of species origin, evolution and specific environmental adaptability.
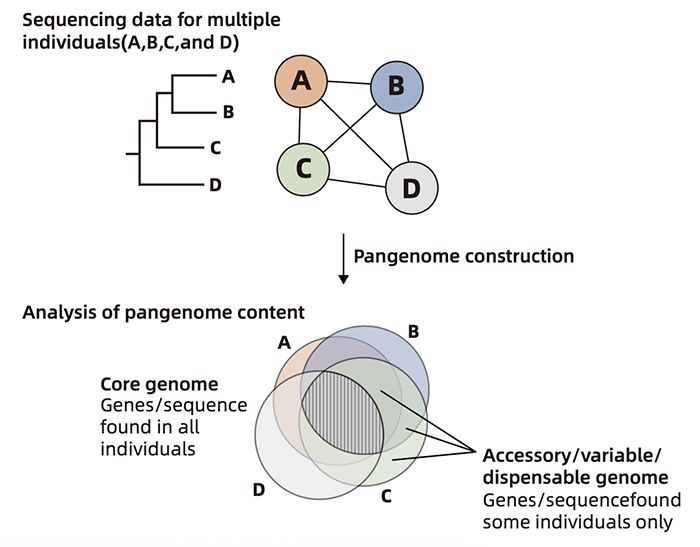
The genome of a single individual cannot fully represent all the genetic information of the species. For example, there is no scab resistance gene PFT in the genome sequence of wheat variety China spring, and there is no flooding resistance gene sub1a in the genome of rice variety Japan Qing. Genome sequence analysis, i.e. pan genome analysis, is carried out for the specific gene information existing in a single sample or part of samples, including core gene, dispensable gene and individual specific gene, which is helpful to fully tap the genetic variation resources and identify the regulatory genes of specific traits of strains. In the future, pan genome will gradually replace the single reference genome and become a "new standard" for the study of animal and plant evolution, selection and gene function.